Article Text
Abstract
Introduction Minority populations in the USA are disproportionately affected by cardiovascular conditions. Reduced responsiveness to clopidogrel among carriers of CYP2C19 variants has been reported in patients with either coronary artery disease (CAD) or acute coronary syndrome (ACS) after the percutaneous coronary intervention (PCI). Previous studies have evaluated CYP2C19 genotyping-guided antiplatelet therapy in selected populations; however, this has yet to be tested among Hispanics. Given the paucity of clinical research on CYP2C19 and antiplatelet clinical outcomes in Hispanics, our study will test the safety and efficacy of a genetic-driven treatment algorithm to guide dual antiplatelet therapy (DAPT) in Caribbean Hispanics.
Methods and analysis This is a multicentre, prospective, non-randomised clinical trial that proposes an assessment of pharmacogenomic-guided DAPT in post-PCI Caribbean Hispanic patients with ACS or CAD. We will recruit 250 patients to be compared with a matched non-concurrent cohort of 250 clopidogrel-treated patients (standard-of-care). Major adverse cardiovascular events (MACEs) such as all-cause death, myocardial infarction (MI), stroke, coronary revascularisation, stent thrombosis and bleedings over 6 months will be the study endpoints. Among the recruited, high-risk patients will be escalated to ticagrelor and low-risk patients will remain on clopidogrel. The primary objective is to determine whether genetic-guided therapy is superior to standard of care. The secondary objective will determine if clopidogrel treatment in low-risk patients is not associated with a higher rate of MACEs compared with escalated antiplatelet therapy in high-risk patients. Patients will be enrolled up to the group’s completion.
Ethics and dissemination Approval was obtained from the Institutional Review Board of the University of Puerto Rico Medical Sciences Campus (protocol # A4070417). The study will be carried out in compliance with the Declaration of Helsinki and International Conference on Harmonization Good Clinical Practice Guidelines. Findings will be published in a peer-reviewed journal and controlled access to experimental data will be available.
Trial registration number NCT03419325; Pre-results.
- coronary heart disease
- coronary intervention
- clinical pharmacology
- genetics
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
Strengths and limitations of this study
This will be the first-ever reported clinical trial on pharmacogenomic-guided dual antiplatelet therapy (DAPT) in Caribbean Hispanics with coronary artery disease/acute coronary syndrome undergoing percutaneous coronary intervention (PCI), leveraging the genetic diversity of this admixed population to improve applicability.
The use of a clinician-oriented, pharmacogenomic-based point-of-care app that allows the integration of patient’s genomics and platelet function into informed clinical decision making provides a real-world study of the implementation and application of algorithmically guided DAPT.
Study population is limited to individuals of Caribbean Hispanic heritage who currently reside in the Commonwealth of Puerto Rico, which might preclude generalisability to other populations.
This study focuses mostly on patients undergoing PCI, and findings may not be directly applicable to other clopidogrel indications.
A possible delay of up to 2 weeks in implementing the genotype-driven treatment algorithm post-PCI may miss some patients who might benefit from earlier intervention.
Introduction
Earlier studies have advanced the case for genotyping to personalise and improve antiplatelet therapy in multiple populations worldwide, but not in any Hispanic populations.1–4 As stated by the updated Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2C19 genotype and clopidogrel therapy, reduced responsiveness to clopidogrel in carriers of CYP2C19 loss-of-function (LOF) alleles has been reported in patients with acute coronary syndrome (ACS) after percutaneous coronary intervention (PCI) for both ST-elevated and non-ST-elevated myocardial infarction (STEMI and NSTEMI).1–6 Double carriers of CYP2C19 LOF alleles had a higher on-clopidogrel myocardial infarction (MI) event rate over a 1-year follow-up period in the French Registry of Acute ST-elevation Myocardial Infarction (FAST-MI) study (adjusted HR=1.98; 95% CI: 1.10 to 3.58).3 Therefore, our trial aims to test the safety and efficacy of a treatment algorithm based on ex vivo pharmacodynamics (PDs) and genetic test results to guide dual antiplatelet therapy (DAPT) in Caribbean Hispanics.
We hypothesise that the use of rapid testing to define high on-treatment platelet reactivity (HTPR) and CYP2C19 LOF allele status to tailor antiplatelet therapy is associated with improved clinical outcomes. Based on prior reports that the clinical implementation of CYP2C19 genotype-guided antiplatelet therapy reduced cardiovascular events after PCI (HR=0.09, p=0.035),7 the rationale behind this study is that switching from clopidogrel to an alternative antiplatelet therapy after PCI in patients with the CYP2C19 LOF genotype will reduce the risk of major adverse cardiovascular events (MACEs). Studies conducted in high-risk settings support the clinical benefit of PD and genetic testing to guide DAPT primarily in patients of Western European ancestry. We will test our hypothesis through a clinical outcome assessment of an antiplatelet treatment algorithm developed using PD and genetic test results within a prospective longitudinal study conducted in 250 Caribbean Hispanics with coronary artery disease (CAD)/ACS requiring DAPT.
Consistent with the goals and mission of the Precision Medicine Initiative and National Institute on Minority Health and Health Disparities (NIMHD), the aim of this research is to enable improved medical cardiovascular outcomes for Caribbean Hispanic patients on antiplatelet therapy through an approach that takes into account individual’s haplotypes, clinical variables, as well as ethno-geographic factors. The ultimate goal is to provide Hispanic patients from Puerto Rico with an equal opportunity to benefit from personalised DAPT, thereby promoting health equity and reducing health disparities.
Methods and analysis
Patient population and study design
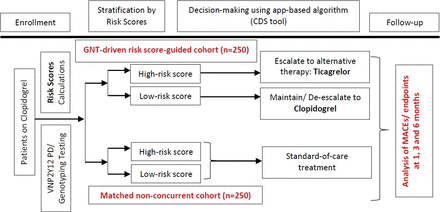
This will be a multicentre, prospective, longitudinal, non-randomised clinical trial study where patients with a high-risk score will be escalated to ticagrelor and patients with a low risk score will be kept or de-escalated to clopidogrel. Patient enrolment is scheduled to begin on 1 September 2020 and expected completion of the study is on 30 June 2022. Prasugrel could be recommended in high-risk patients with any contraindication to ticagrelor. The genetic-driven risk score-guided subcohort (n=250) will be compared with a matched non-concurrent subcohort of clopidogrel-treated patients (n=250). The non-concurrent subcohort will be derived from a cardiovascular genome-wide association study (GWAS) currently being conducted as a discovery phase of the proposal to identify other genetic variants that may be relevant to clopidogrel treatment failure (n=750, to be recruited between January 2018 and June 2020). From this discovery group, we will include 250 participants matched by age, gender, risk scores and other relevant covariates to those finally enrolled in the guided subcohort. Study protocol (A4070417, Title: Adopting a Precision Medicine Paradigm in Puerto Rico: leveraging ancestral diversity to identify predictors of Clopidogrel response in Caribbean Hispanics) was registered at the US National Institutes of Health website and is currently ongoing (http://www. clinicaltrial.gov).
Inclusion and exclusion criteria (table 1) are minimal with the goal of gathering an ‘all comers’ population representative of real-world clinical practice—regardless of platelet reactivity status or presence of CYP2C19 polymorphisms, in order to better assess PD profiles and clinical outcomes. Patients with contraindications/precautions for both ticagrelor and prasugrel will be excluded from further analyses.
The criteria for inclusion/exclusion in the study are as follows
After signing informed consent, patients in the genotype-driven risk score-guided subcohort will undergo rapid ex vivo platelet and genetic testing (ie, expected turnaround time of approximately 3–5 days) and a blood draw for additional assessments and sample storage for future genetic analysis (broad consent obtained). Patients in this subcohort will be further categorised into two (2) groups based on test results and the corresponding risk score calculations:
Patients with high-risk scores.
Patients with low-risk scores.
An optimal antiplatelet therapy will be recommended postintervention to the primary cardiologists, according to the category in which the patient is identified, as described in the treatment algorithm outlined in figure 1. Recommendations for optimising therapy will be available through a custom mobile application (app) within 2 weeks of hospital discharge. To ensure full engagement, participating clinicians will be educated on proper use of the mobile app. This is a clinician-oriented, pharmacogenomics-based point-of-care (POC) app that was developed in-house by our bioinformatics and technology (IT) team to allow the integration of patient’s genomics and PD into informed clinical decision making (ie, clinical decision support tool) so that clinicians may easily apply algorithmically guided DAPT plans in real-time, on-demand, in clinical settings.
Experimental design. CDS, clinical decision support tool; GNT, genotype; MACEs, major adverse cardiovascular events; PD, pharmacodynamic.
Changes in DAPT (ie, escalations/de-escalations of therapies) will always be at clinician’s discretion. In particular, alternative therapy with ticagrelor (according to specific contraindications and precautions for this agent8) will be strongly recommended for high-risk score patients, whereas de-escalating or maintaining clopidogrel for low-risk score patients will be a moderate recommendation. Treatment strategies and clinical outcomes will be evaluated up to 6 months. To minimise dropouts, we will offer transportation to participants, provide calendars and pre-appointment reminders, keep contact by periodic phone calls and build trusting relationships with participants, their families and primary care physicians.
Blood samples (20 mL) for genetic testing, ex vivo residual platelet function analysis and storage at −70°C will be collected from patients’ peripheral veins at a single time point (day 0), within a week of first clopidogrel dose. Blood will be collected into EDTA and sodium citrate (3.2%) tubes. Platelet reactivity will be measured using the VerifyNow P2Y12 assay (Accumetrics, San Diego, California, USA), following the manufacturer’s instructions (ie, assessed within 4 hours after blood withdrawal but discarding the first volume of 3 mL of blood). The results will be expressed as P2Y12 reaction units (PRU). HTPR is defined as ≥208 PRU, based on prior reports.9–11 DNA will be extracted using standard methods, and CYP2C19 genotypes will be determined by TaqMan SNP Genotyping assay methodology for StepOnePlus Real-Time PCR system, according to standard protocols. Additional genotype assessments may include B4GALT2, CES1, PON1 and ABCB1, as these are potentially related to platelet activation, metabolism and absorption of P2Y12 receptor inhibitors.
Risk score calculation
Patients will be given 2 points for HTPR or genetically inferred CYP2C19 poor metaboliser status (ie, mainly due to the presence of the CYP2C19*2 allele), 1 point for CYP2C19 intermediate metaboliser phenotype and 0.5 point each for the following: presence of type 2 diabetes mellitus diagnose, adjoined length of stent(s) >30 mm, a left ventricular ejection fraction <30% or any variant allele in loci of GWAS significance identified in the discovery phase.12 On the contrary, patients will be subtracted 0.5 points per each PON1 p.Q192R allele or haematocrit (Hct, >50%) changes above normal range. Patients with a risk score of ≥2 will be allocated to the high-risk score group, and clinicians will be advised to switch/escalate this group of patients to ticagrelor in the genotype-driven risk score-guided cohort; patients with a risk score <2 will be classified as low risk and a recommendation will be made for those in the genotype-driven risk score-guided cohort to be continued on clopidogrel treatment (or de-escalated from ticagrelor/prasugrel to clopidogrel).12 The risk scores will be calculated using the same methodology for the non-concurrent control group, although in this group clinicians were not aware of patient risk scores and therefore did not receive any recommendations with regard to escalation/de-escalation of therapy.
Study endpoints and statistical analysis
Clinical outcomes will be recorded to evaluate changes in antiplatelet treatment strategy. Follow-up will be performed by research staff at 1, 3 and 6 months through medical records reviews, patient’s clinical visits and phone calls. We believe this follow-up period is appropriate because most potentially preventable recurrent events occur within the first 6 months of treatment.13–16 Adherence to the prescribed medications will be assessed by proportion of days covered (≥80% considered compliant)17 18 and interview with a Standardised Medication Assessment Questionnaire (ie, ≥3 pts on the original four-item Morisky, Green and Levine (MGL MAQ) Medication Adherence Scale in the public domain).19 Treatment patterns will be evaluated according to the assigned subcohort and clinical outcomes will be evaluated.
The primary endpoint (ie, MACEs) will be the composite of all-cause death, non-fatal non-procedural MI (according to the universal definition20), nonfatal ischaemic stroke, coronary revascularisation and non-fatal non-procedural definite stent thrombosis during 6 months of follow-up. An event will be considered procedural when occurring within 24 hours of PCI and, therefore, excluded from the corresponding endpoint.
Secondary endpoints will include cardiovascular death and each individual MACE endpoint in the above composite. Major and minor bleeding events will also be analysed as secondary endpoints, with bleeding defined by the Bleeding Academic Research Consortium and Thrombolysis in Myocardial Infarction (TIMI) bleeding criteria.21 Efficacy and safety endpoints will be evaluated by blinded event adjudication by our study clinicians, relying on original source data. In the event of reporting a serious adverse event, antiplatelet treatment will be managed according to standard of care. Independent predictors of HTPR in our study cohort will be evaluated taking into account clinical and genetic data. Clinical outcomes will be compared according to received treatments and computed risk scores based on HTPR/LOF status. Since data on HTPR/LOF status (ie, ex vivo platelet function and genetic test results) will also be available for patients in the non-concurrent subcohort, risk scores will be estimated in this group as well and used for outcome comparisons accordingly, but not for treatment decision making. By design, this non-concurrent subcohort will follow the standard-of-care therapy as recommended by current guidelines and medical judgements.
The guided and non-concurrent subcohorts will be compared, but comparisons will also be made based on stratification by high-risk and low-risk score, both within and between subcohorts. First, we hypothesise that the rate of MACEs in patients with low-risk scores who were maintained/de-escalated to clopidogrel will be non-inferior to that achieved by an alternative antiplatelet therapy (ie, ticagrelor) in the high-risk score subgroup. This is intended to demonstrate whether clopidogrel is non-inferior to therapy with ticagrelor when used only in non-resistant patients. In addition, we also hypothesise that the rate of MACEs in a genotype-driven risk score-guided subcohort is lower than that in the non-concurrent subcohort. This will demonstrate whether an antiplatelet therapy guided by genotypes (CPIC guideline) and risk scores is superior to the current standard of care. Based on preliminary data and prior literature reports, we will assume a primary outcome rate (MACEs) of not less than 3% in the study cohort.22–24 Likewise, we anticipate a prevalence of HTPR and CYP2C19 LOF alleles combined of approximately 30%.10 11
Sample size and power calculation
A sample size of 500 subjects (250 per subcohort) will be included in our study. Logistic regression power analysis (power log command) was used in Stata V.16 to determine the detectable effect size (odds ratios) with 80% statistical power and an alpha of 0.017 (Bonferroni adjustment), based on different assumptions for MACE incidence in the reference groups. All calculations assumed that 30% of the subjects within the genotype-guided subcohort (n=75) will present high-risk scores and will be escalated to ticagrelor.10 As presented in table 2, the detectable OR estimates for pairwise tests varied, starting from 1.9 for comparing patients with therapy based on current guidelines versus low-risk patients maintained/de-escalated to clopidogrel, 2.5 for low-risk patients maintained/de-escalated to clopidogrel versus high-risk patients switched/escalated to ticagrelor and 2.4 for patients with therapy based on current guidelines versus high-risk patients switched/escalated to ticagrelor.
Detectable effect size with 80% statistical power and alpha of 0.017 according to group comparison and incidence of MACE among the reference group
The upper bound of the CI for the true difference of rates (%) must not exceed a clinically meaningful non-inferiority margin, δ. If there is truly no difference between both subgroups, then 500 patients (250 per group) will suffice to be 80% sure that the upper limit of a one-sided 95% CI will exclude a difference in favour of the alternative subgroup of more than 5.9%. This sample size will also allow for a reasonable dropout rate (or rate of invalid results) of 5%. To minimise potential bias, both intention-to-treat and per-protocol approaches will be used.
Statistical analysis plan
Propensity scores will be calculated as a conditional probability of being in the ticagrelor treatment group using logistic regression models. Inverse probability of treatment (IPT) weighting will be determined using the propensity scores in order to obtain estimates of average treatment effects on subjects. To avoid a high variability of the estimated treatment effect, weights will be trimmed at the 3rd/97th percentile; trimmed cut-off point may change according to the tail properties and/or the treatment prevalence. To assess balance of covariates in the propensity scores, standardised differences will be compared between treatment groups in the weighted samples. In addition, distribution of continuous covariates will be assessed through side-by-size boxplots and empirical cumulative distribution functions.
Descriptive statistics (eg, percentages for categorical variables; means±SD, medians and interquartile ranges for continuous variables) will be used to characterise participants in the study. Patient background characteristics will be compared between treatment groups (ie, (1) high-risk patients switched/escalated to ticagrelor, (2) low-risk patients maintained/de-escalated to clopidogrel and (3) patients with therapy based on current guidelines/medical judgement) using χ2 (or Fisher’s exact test) for categorical variables and analysis of variance/Kruskal-Wallis test for continuous variables (depending on the results of the Shapiro-Wilk normality and Bartlett’s homoscedasticity tests).
The primary endpoint (incidence of MACEs) will be assessed to determine if clopidogrel is non-inferior to therapy with ticagrelor when used only in non-resistant patients. This will be achieved by comparing the incidence of MACEs between treatments using logistic regression models. IPT-weighted ORs along with their 95% CIs will be reported.
Furthermore, Kaplan-Meier analysis will be used to estimate the survival function (MACE free) over a 6-month period; log rank test will be used to determine differences in survival distributions between treatments and subcohort groups. IPT-weighted Cox proportional hazards models will also be performed to regress survival time according to treatments and subcohort groups. All executed tests for primary endpoints will also be performed for the secondary endpoints. Significance level of statistical tests will be adjusted for multiple comparisons if needed. Data analyses will be performed using Stata V.16.
Ethics and dissemination
The study was approved by the local Institutional Review Board (IRB) ethics committee (Federal-wide Assurance #00005561) of the University of Puerto Rico Medical Sciences Campus (UPR-MSC, San Juan, Puerto Rico, USA), under approval #A4070417. This study will be carried out in compliance with the Declaration of Helsinki and International Conference on Harmonization Good Clinical Practice Guidelines. A waiver for authorisation to access medical records of potential participants was granted in order to identify in advance those individuals who meet inclusion criteria. Each study participant will receive a verbal and written description of the study. Subjects are also advised of the voluntary nature of participation and of their right to withdraw freely from the study at any time without prejudice for their future care as well as to require that information about them be removed from data analysis. A fully bilingual study coordinator will carry out the recruitment using culturally appropriate means of communication. A clause of broad consent for future research is included as a part of the informed consent form (online supplementary material S1). Participants will be also asked to sign this portion of the informed consent in order to explicitly obtain authorisation for collection and storage of their deidentified biological specimens.
Supplemental material
All applicable ethical, legal and social implications of genetic testing and interpretation, data collection and specimen storage in this study will also be discussed with participants as part of the consent process. All genetic and clinically relevant data arising from this study will be recorded, curated and entered into a standardised Health Insurance Portability and Accountability Act (HIPAA)-compliant database system for analysis and deposit. As our population includes only Hispanic patients, all project documents, including the consent forms, will be available in culturally appropriate Spanish language. If a participant’s reading skills are limited, all materials will be presented verbally in Spanish or English, as indicated. The consent process takes into account the questions whether and when investigators will re-contact participants. A manuscript presenting and discussing all relevant findings from this study protocol as well as results of covariate analyses will be submitted and published in a peer-reviewed journal. On completion of the trial and publication of the results, a request for experimental data can be submitted to the principal investigator at the University of Puerto Rico, Medical Sciences Campus in San Juan, Puerto Rico, USA.
Data and sample management
Proper safeguards against any potential violation of privacy or breach of confidentiality will be provided. In this protocol, confidentiality of collected data will be preserved by using a code number to each participant (ie, a seven-digit unique study identification number). Codes will neither contain nor scramble letters or numbers that might be linked to individually identifiable information from patients enrolled in the study. No individual personal information will be disclosed. Cautions will be observed to protect and avoid unnecessary disclosure of any research-related health information arisen during this study. Subjects’ identities and numbers will only be accessible to designated investigators and all data will be treated in a non-nominal manner. All documentation related to study as well as a password-protected laptop containing encrypted files from participants will be kept in locked drawers. Access to the building is restricted. All staff entrances are secured with locked, coded doors and changed regularly. The computing system is protected from outside access by a two-tiered firewall system: the Linux Server running Net filter is backed-up by the Windows Server running Microsoft Internet Security and Acceleration Server (ISA). Both servers maintain logs and generate reports of access attempts, which are reviewed by the Network Administrator. All data sent outside the study facility will be encrypted and have tracking documents attached to ensure ‘chain of custody’.
Fresh whole blood samples collected in vacutainers will be immediately coded using the corresponding patient’s study number printed on each specimen tube label so that the sample will not directly identify the patient. Deidentified DNA specimens will solely be used for genetic analyses related to this study, except for those individuals consenting for DNA storage in a local repository (biobank), where they will be stored frozen in Eppendorf tubes. Otherwise, it will not be used to serve any other purpose than that specifically defined in the study protocol. All blood samples and DNA specimens of individuals not consenting for the repository will be properly disposed after processed, following standard procedures for safety disposal of biohazard materials. All investigators will sign confidentiality agreements and have completed the appropriate data security training. The study data might be reviewed by the ethics committee overseeing the research. After completing the protocol, data will be stored as long as it is required by institutional policies (usually for a maximum period of 5 years) and then disposed following an accepted deidentification procedure (overwriting or degaussing). Data and DNA specimens stored in biobank can be used after the study if a broad consent for future studies is properly obtained from participants.
Patient and public involvement
Through our Community Engagement Core Unit of the Center for Collaborative Research in Health Disparities (RCMI-CCRHD) at the UPR MSC, patients and the public were partially involved in the design and dissemination plans of our research by discussing this protocol with community leaders, members of local health organisations (eg, American Heart Association), patients and patient advocates who provided feedback on how to conduct the study as well as the best way to disseminate findings from this experimental protocol.
Discussion
In this trial, determinants of decreased antiplatelet drug effects will be investigated and an ethno-specific treatment algorithm for risk stratification (see Risk score calculation) will be generated and implemented by clinicians to tailor antiplatelet therapy in Caribbean Hispanics with CAD/ACS undergoing PCI on DAPT. The risk score was developed by extracting key clinical, genetics (ie, CYP2C19*2 allele) and lifestyle indicators from our previous studies10 11 as well as from those by others (ie, the POPular Risk Score)12 on how critical factors influence ultimate outcomes in patients from a similar study cohort (table 3). The latter report describes how points are given in the risk score computation, which will be used the same way in the proposed protocol. The POPular Risk Score, which is based on platelet reactivity (VerifyNow P2Y12 assay), CYP2C19 genotyping and clinical risk factors, was developed for the selective intensification of P2Y12 inhibitor treatment with prasugrel instead of clopidogrel in patients undergoing non-urgent PCI with stent implantation.12 The use of this score in clinical practice was found to be associated with a reduction in thrombotic events without an increase in bleeding events12; therefore, we decided to adopt a similar approach in our study protocol.
Factors used to calculate the risk score
Since this treatment algorithm (figure 1 and online supplementary material S2A-B) is based on the unique genetic backgrounds of Caribbean Hispanics, a highly admixed population that is ethnically distinct to other populations from previous studies, it is deemed an ethno-specific tool for supporting treatment decisions and offering guidance on clinically actionable recommendations in Caribbean Hispanics. We firmly believe that tailoring antiplatelet therapy (maintaining clopidogrel or switching to prasugrel or ticagrelor) according to an algorithm that incorporates clinical characteristics, platelet reactivity and genotyping will be associated with improved clinical outcomes.25 26 In particular, we expect that Caribbean Hispanics with HTPR to clopidogrel or CYP2C19 LOF switched to ticagrelor will have better outcomes compared with those maintained on clopidogrel, and similar outcomes compared with patients without HTPR or CYP2C19 LOF. Prior investigations are limited by the low-risk profile of the study populations, under-representation of minorities and non-white ethnicities, narrow use of more effective antiplatelet therapies (ie, prasugrel and ticagrelor) and lack of clinical characteristics consideration in directing the therapy.27–30 The study will include a high-risk patient subcohort of Caribbean Hispanics, risk-stratified using both genetic and PD testing, and adopt ticagrelor therapy.
Supplemental material
We anticipate that the treatment algorithm, based on rapid ex vivo platelet function and genetic testing, will lead clinicians to implement personalised DAPT in Caribbean Hispanics with the ultimate goal to improve adverse clinical outcomes. A similar approach has proved valid in other populations.7 Other factors may be associated with impaired platelet inhibition and affect clinical outcomes, such as clinical variables, which will then be implemented in our flexible app-based algorithm and used to build models able to better predict clinical outcomes in patients requiring DAPT.
The non-concurrent subcohort as a control group for this study was chosen for several reasons. First, although the use of a pharmacogenetic-driven algorithm to guide DAPT has yet to be studied in a Caribbean Hispanic population, CPIC guidelines have already established actionable CYP2C19 genotype recommendations. However, CYP2C19 genotyping is not yet common in Puerto Rican Institutions. Given the existing evidence that supports the use of genotyping results to guide therapy, our clinicians were not comfortable having a control group that was genotyped, but then blinded from results and an intervention that could have beneficial clinical effects. The use of a non-concurrent subcohort of patients who have been genotyped and observed clinically before the pharmacogenetic-driven algorithm was implemented provides an excellent alternative for a control group that does not necessitate the withholding of a now accessible clinical intervention with probable clinical benefit. As is often the case when translating pharmacogenomic results into clinical practice, findings require validation in large, ‘real-life’ populations or population-based cohorts with individuals from multiple races/ethnicities. While randomised clinical trials are useful in early discovery phases as subjects tend to have similar attributes and adherence to medications tends to be higher than normal, leading to more clear results that however lack generalisability. Leveraging the genetic diversity of the real-life, admixed population in this study will improve the applicability of our findings.
We propose a non-inferiority design to show that the primary outcome rate for low-risk patients on clopidogrel is not substantially higher than the corresponding rate for high-risk patients on an alternative antiplatelet therapy (ie, ticagrelor). For this study, we have chosen a composite endpoint (MACE), as is common in previous work investigating pharmacogenetic determinants to clopidogrel response.1–4 7 The rationale for the use of MACE as the primary endpoint is that all of the individual components of MACE are common clinical sequelae in patients undergoing treatment with DAPT, and the occurrence of any one of these components is considered resistance to or failure of treatment. This non-randomised clinical trial study will also gather clinical non-genetic data, as well as additional genetic tests not directly included in the risk score calculation by interrogating other candidate pharmacogenes (ie, B4GALT2, CES1 and ABCB1)31–33 and any other top GWAS signals we have previously identified in this study population. Conversely, we plan to perform additional assessments to test population-specific factors (ie, genetic and non-genetic relevant biological variables) that are independently associated with either HTPR or low on-treatment platelet reactivity in Caribbean Hispanics on clopidogrel with CAD/ACS. Studies have shown an independent association of race with HTPR even after adjusting for CYP2C19 phenotype and that genetic factors other than CYP2C19 polymorphisms are associated with outcomes in patients with CAD/ACS.34 This suggests that differences in risk factor distribution and other genetic variants may be associated with increased thrombogenicity in our study population. Platelets respond to many factors such as smoking and other medications.35 36 A significant proportion of carriers of the loss-of-function CYP2C19 allele do not have high post-clopidogrel residual platelet reactivity, and high residual reactivity is present in patients with wild-type alleles.5 Functional tests of platelet reactivity therefore furnish a dynamic result, whereas the genotyping result is innate to the patient.34 37 38 Ultimately, we expect that our clinical approach that incorporates clinical characteristics, platelet reactivity and genotyping will be associated with improved clinical outcomes.
A limitation of this study is the possible delay of up to 2 weeks in implementing the genotype-driven treatment algorithm post-PCI, as studies have proven that incidences of stent thrombosis are highest within the first 4 weeks post stenting.9 23 Although we recognise that a POC test capable of providing on the spot genotype results prior to the patient’s discharge after the PCI is ideal, it is beyond our reach at present. Since several PCIs will be performed on elective basis, many patients will have less than 24 hours of hospital stay; therefore, implementing our algorithm during the admission would be extremely challenging. However, we expect to fully implement the genetic-driven treatment algorithm within the first week post-PCI for the vast majority of our patients.
Acknowledgments
We would like to thank the patients for voluntarily participating in this study protocol. A special acknowledgement to the PRCTRC’s Research Design and Biostatistics Core service for helping us with study design and sample size calculations.
References
Footnotes
DFH-S and KM are joint first authors.
DFH-S and KM contributed equally.
Contributors DFH-S, KM, FM-M and JD conceived the study hypothesis, designed and wrote the trial protocol. LG-S and SR-T performed the power/sample size calculations and statistical design. HN, AFG, HJN, GR and SAS provided revision of the proposed protocol and participated in the write-up of the manuscript. All the authors contributed to the feasibility of the study, revised, edited and approved the final version of this manuscript.
Funding This work was supported by the Center for Collaborative Research in Health Disparities (RCMI-CCRHD) grant #U54 MD007600-31 from NIMHD, National Institutes of Health (NIH). PRCTRC’s Research Design and Biostatistics Core service is supported by the NIMHD and the National Institute of Allergy and Infectious Diseases (NIAID) under award # U54MD007587. The funding sources for this trial had no role in the design of the study and will not have any role in the performance of the research.
Disclaimer The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Competing interests None declared.
Patient and public involvement Patients and/or the public were involved in the design, or conduct, or reporting, or dissemination plans of this research. Refer to the Methods section for further details.
Patient consent for publication Not required.
Ethics approval Institutional Review Board (Federal-wide Assurance Number 00005561) of the University of Puerto Rico Medical Sciences Campus (UPR-MSC, San Juan, PR, USA).
Provenance and peer review Not commissioned; externally peer reviewed.