Article Text
Abstract
Objective Quantitatively evaluate the quality of data underlying real-world evidence (RWE) in heart failure (HF).
Design Retrospective comparison of accuracy in identifying patients with HF and phenotypic information was made using traditional (ie, structured query language applied to structured electronic health record (EHR) data) and advanced (ie, artificial intelligence (AI) applied to unstructured EHR data) RWE approaches. The performance of each approach was measured by the harmonic mean of precision and recall (F1 score) using manual annotation of medical records as a reference standard.
Setting EHR data from a large academic healthcare system in North America between 2015 and 2019, with an expected catchment of approximately 5 00 000 patients.
Population 4288 encounters for 1155 patients aged 18–85 years, with 472 patients identified as having HF.
Outcome measures HF and associated concepts, such as comorbidities, left ventricular ejection fraction, and selected medications.
Results The average F1 scores across 19 HF-specific concepts were 49.0% and 94.1% for the traditional and advanced approaches, respectively (p<0.001 for all concepts with available data). The absolute difference in F1 score between approaches was 45.1% (98.1% relative increase in F1 score using the advanced approach). The advanced approach achieved superior F1 scores for HF presence, phenotype and associated comorbidities. Some phenotypes, such as HF with preserved ejection fraction, revealed dramatic differences in extraction accuracy based on technology applied, with a 4.9% F1 score when using natural language processing (NLP) alone and a 91.0% F1 score when using NLP plus AI-based inference.
Conclusions A traditional RWE generation approach resulted in low data quality in patients with HF. While an advanced approach demonstrated high accuracy, the results varied dramatically based on extraction techniques. For future studies, advanced approaches and accuracy measurement may be required to ensure data are fit-for-purpose.
- heart failure
- cardiac epidemiology
- health informatics
Data availability statement
No data are available.
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
STRENGTHS AND LIMITATIONS OF THIS STUDY
Using real-world evidence (RWE) for patients with heart failure (HF) requires demonstrating that the data source and technologies result in accurate data.
Natural language processing alone lacked context from the longitudinal record, limiting phenotype identification and study validity.
Findings suggest that advanced methods can enable high-validity RWE for patients with HF.
The use of data from a single healthcare system may limit generalisability to other populations.
Introduction
Heart failure (HF) is a major public health problem with significant associated morbidity, mortality and cost.1 2 Despite the availability of novel drugs and devices, morbidity and mortality in HF rivals many malignancies, with a 5-year survival rate as low as 50%.3–8 Randomised controlled trials (RCTs) have traditionally been used to assess the safety and efficacy of new therapies and represent a cornerstone for regulatory approval. However, RCTs are frequently conducted in highly selected populations, typically younger, healthier and less diverse than patients treated in clinical practice. Furthermore, such trials often include patients with an established HF diagnosis, receiving guideline-directed medical therapy at tertiary centres, and may not represent the broader population with HF. Because HF is a clinically heterogeneous syndrome with numerous aetiologies and phenotypes, studying this population can be particularly difficult.
Real-world evidence (RWE) has held promise as a potential means to assess therapeutic benefit outside of clinical trials, with sufficient power to characterise therapeutic impact in HF subgroups. Accordingly, RWE can complement RCTs, extending the findings to patient populations that may have been excluded from or insufficiently enrolled in pivotal trials. To accelerate these and similar precision medicine goals, the 21st Century Cures Act was passed in 2016, which required the United States Food and Drug Administration to develop guidance supporting the use of RWE in new drug indications and postmarketing surveillance.9 In addition, payors have increasingly utilised RWE to inform reimbursement decisions and are increasingly demanding credible evidence.10
Not surprisingly, the quality of RWE hinges on how well real-world data are collected, processed11 and used to inform study questions. Such is the case in HF, where accurate identification of patients in administrative and other structured datasets is an ongoing focus.12–14 Traditional methods of identifying patients with HF rely on querying diagnosis codes and structured data in the electronic health record (EHR) or medical claims. Conversely, artificial intelligence (AI) applied to unstructured data represents a novel method of analysing the medical record. Because of the importance of data reliability in RWE and the potential to use unstructured data to achieve data enrichment,15 we sought to compare the accuracy achieved by traditional RWE methods versus advanced AI approaches in identifying a range of HF-specific data elements from the medical record.
Methods
The study design is outlined in figure 1. Varied data sources and applied technologies were used to assess data reliability in patients with risk factors for HF. Leveraging manual chart abstraction as the reference standard, comparisons were made between the two methods. The first method used structured EHR data (eg, diagnosis codes and problem lists) and standard query techniques, defined as the ‘traditional approach’. The second used unstructured EHR data (eg, narratives from primary care and specialty notes) and AI techniques, described as the ‘advanced approach’ (figure 1). The primary objective was measurement of the accuracy of identified HF-specific elements using traditional and advanced approaches. We hypothesised that the advanced approach would better identify key HF-specific elements than the traditional approach. Data were deidentified before study initiation, and the study was determined not to be human subjects research. Both natural language processing (NLP) and machine-learning inference technologies used in the advanced approach were provided by Verantos (Menlo Park, California, USA). The core of AI is a deterministic NLP layer. This layer is built on top of the GATE NLP architecture.16 The architecture is used to construct a flexible pipeline for processing incoming text against English language syntactical rules augmented with a lexicon based on a clinical vocabulary. The AI-based inference was applied during data processing. Millions of machine-learning and manually curated associations enable disambiguation and identification of clinically relevant concepts. As an example of AI-based inference, a patient with HF on the problem list and a narrative encounter describing ‘EF 60%’ would not be interpreted by NLP as having HF with preserved ejection fraction (HFpEF) since the text does not have sufficient information to identify this condition. On the other hand, AI-based inference would infer HFpEF based on disparate information in the record.
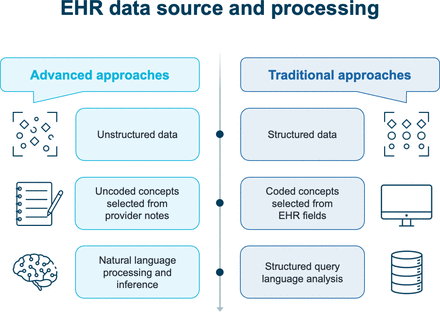
EHR data source and processing. EHR, electronic health record.
EHR data source and processing
EHR data from primary care encounters between 2011 and 2018 were deidentified and securely transferred to a cloud-based server for analysis. The dataset consisted of both structured data (eg, medical conditions, procedures performed, medications and problem lists) and unstructured data (eg, narrative notes from primary care providers and specialists, telephone visits, and other narrative text) (figure 2).
Comparison of traditional and advanced real-world evidence approaches. EHR, electronic health record.
As the study aimed to test the accuracy of different RWE approaches and not treatment effectiveness, the cohort was enriched for patients with suspected HF based on comorbidities and medications. Specifically, the following filters were applied: records containing both narrative and structured components; narrative length 1000 characters or more; and at least one of the following problems or medications in structured or unstructured data—myocardial infarction, congestive HF or carvedilol (figure 1).
A prespecified set of clinical concepts pertinent to patients with HF was extracted using traditional and advanced techniques (table 1). Problem lists were mapped to Systematised Nomenclature of Medicine (SNOMED) ontology, and unadjudicated claims were mapped to International Classification of Diseases (ICD)-10 codes. Standard sets of individual codes were used to represent each concept. With the advanced approach, inference incorporating pattern recognition was utilised to identify potentially missing or ignored concepts within the text (eg, HF being likely in patients with dyspnoea and pitting oedema on a diuretic). Specifically, no narrative coding took place before the AI algorithm was used; instead, it was applied directly to the narrative text and then mapped by the algorithm to the SNOMED ontology. Next, manual chart abstraction using the same SNOMED code set was used as a reference to assess the accuracy of the coding by the AI algorithm. Engineers were blinded to validation data and its corresponding chart abstraction.
Prespecified HF-specific concepts extracted from the electronic health record
Study end points and statistical analysis
The primary endpoint was the F1 score for traditional and advanced approaches. The F1 score is an accuracy measure that combines recall and precision; more specifically, it is the weighted harmonic mean of these two measures. Secondary endpoints were recall (ie, the proportion of patients correctly identified as having the condition, akin to sensitivity) and precision (ie, the proportion of patients with HF and its subtypes correctly identified divided by the total number of patients identified in each cohort akin to positive predictive value)17 18 for the traditional and advanced approaches. The reference standard used to evaluate accuracy of the traditional and advanced approaches was manual chart abstraction. For each encounter, two independent clinical annotators labelled each concept and all metadata for that concept. Annotators were blinded to each other’s annotations, and inter-rater agreement was measured by Cohen’s kappa score. Further description of the reference standard methodology is provided in the online supplemental material. Results were summarised using descriptive statistics, and percentages were calculated for categorical variables. Differences in F1 scores between traditional and advanced approaches were analysed using the χ2 test; associated p-values were reported.
Supplemental material
Patient and public involvement
Data were deidentified before study initiation, and the study was determined not to be human subjects research. As a result, no patients were recruited for study participation. The research question and study goal of highlighting methods for improving RWE use were driven by recognition that improvements in use of RWE to inform new drug indications, postmarketing surveillance, and reimbursement decisions would ultimately result in patient benefit.
Results
A total of 4288 encounters for 1155 patients were examined, of which 472 patients with HF were identified. Of these, 382 had HF with reduced ejection fraction (HFrEF), 35 had HF with mildly reduced ejection fraction (HFmrEF) and 55 had HFpEF. The reference standard Cohen’s kappa score was 0.95, suggesting high validity.
Online supplemental table 1 reports the F1 score, recall and precision results achieved with both approaches. Figure 3 graphically presents F1 scores for HF diagnoses and figure 4 includes F1 scores for symptoms, medications and comorbid conditions. Overall, accuracy was significantly greater for the advanced approach (AI applied to unstructured EHR data) than for the traditional approach (structured query language applied to structured EHR data) (online supplemental table 1; figures 3 and 4), with an absolute difference of 45.1%.
Supplemental material
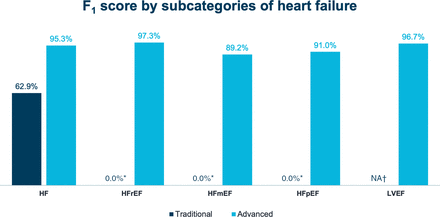
F1 scores for HF diagnoses. 0% reflects a measured value and indicates the availability of the diagnosis code in the EHR dropdown versus N/A, which refers to a diagnosis without available code in the relevant codeset. *F1-score could not be calculated due to lack of data for precision. †Structured data recall is not applicable for ejection fraction because no code was available within the problem list. HF, heart failure; HFmrEF, heart failure with mildly-reduced ejection fraction; HFpEF, heart failure with preserved ejection fraction; HFrEF, heart failure with reduced ejection fraction; LVEF, left ventricular ejection fraction; N/A, not applicable.
F1 scores for (A) symptoms, (B) comorbid conditions and (C) medications. *F1 score could not be calculated due to a lack of data for precision.
With the traditional approach, recall for any HF diagnosis was 46.9% (ie, 53.1% of patients with HF were missed entirely) and precision was 95.4%, resulting in an F1 score of 62.9% (p<0.001). In contrast, with the advanced approach, recall for any HF diagnosis was 96.0% and precision was 94.7%, resulting in an F1 score of 95.3% (p<0.001 when F1 scores for the two approaches were compared) (online supplemental table 1; figure 3). Among HF phenotypes, recall with the advanced approach was highest with HFrEF, followed by HFpEF and HFmrEF; precision was 100% for all phenotypes. With the traditional approach, F1 scores could not be calculated for HFrEF, HFmrEF and HFpEF because only less granular HF codes were used (online supplemental table 1).
Accuracy in identifying left ventricular ejection fraction (LVEF) was similarly high with the advanced approach, with an F1 score of 96.7%. Data could not be extracted for LVEF with the traditional approach because no such codes were available within the EHR, nor did a mechanism to encode LVEF within the problem list or unadjudicated claims exist (online supplemental table 1; figure 3).
Accurate identification of HF symptoms was greater with the advanced approach (p<0.001) (online supplemental table 1; figure 4A). Although identification of commonly prescribed HF medications was high with both approaches (online supplemental table 1; figure 4B), identification of cardiovascular comorbidities was higher in all cases with the advanced approach (p<0.001) (online supplemental table 1; figure 4C).
Data concept extraction with the advanced approach greatly depended on the technology used. For example, NLP, which ends at the sentence boundary, was only able to identify HFpEF with an F1 score of 4.9% because ‘HFpEF’ or ‘heart failure with preserved ejection fraction’ was rarely written. Conversely, inference, which can find related items from the longitudinal record, was able to identify both ‘HF’ and ‘normal ejection fraction’ as separate annotations for HFpEF with an F1 score of 91.0% (online supplemental table 1; figure 3).
Discussion
The utilisation of RWE has grown substantially in recent years, driven in part by its perceived value by clinicians, regulators and payors, particularly in light of the limitations of trial populations.19 As RWE is increasingly used to refine care standards through clinical, regulatory and reimbursement pathways, its accuracy has come under increased scrutiny. This is particularly important for complex medical conditions, such as HF.20 Accordingly, in this analysis, chart abstraction was used to quantitatively evaluate traditional and advanced approaches to define HF-specific data elements. This enabled rigorous evaluation of whether commonly used techniques are sufficiently accurate for observational studies, comparative effectiveness research and post-approval safety studies.
In this study, (1) the use of an advanced, AI-based approach consistently identified HF phenotypes (ie, HFrEF, HFmrEF and HFpEF) more accurately than a traditional approach; (2) common HF symptoms and comorbid conditions were consistently and accurately identified using an advanced approach; and (3) medications for HF were accurately identified using both advanced and traditional approaches. While studies have previously leveraged an AI-based approach to identify patients with HF,21–24 the findings presented here highlight the discrepancy between traditional EHR query methods and an AI-based approach standardised against a manual reference. Given that the accuracy of the dataset and appropriateness of the applied technology are not tested in many RWE studies, there is a high potential for error.25–28 The current findings highlight this while also reinforcing the impact that specific AI technologies (eg, NLP vs NLP plus inference) can have on phenotype generation and study validity.
Accurate phenotyping is paramount in any RWE study that includes patients with HF. With varying aetiologies and multiple phenotypes, HF is a clinically diverse syndrome, with outcomes that may vary between and even within subgroups.29 30 In addition, patients with HF may have different trajectories, highlighting some of the limitations of using structured data. For example, LVEF may fluctuate throughout a patient’s disease course, with some patients experiencing recovery of their LVEF with the use of guideline-directed medical therapy. Accordingly, accurate phenotyping of patients with HF usually requires the incorporation of data that crosses clinical encounters. In addition, although symptoms are an essential reflection of clinical status, they are poorly captured in structured data. Suboptimal recognition of comorbidities like valvular heart disease can also impact disease trajectory and risk for future cardiovascular events.
The findings presented here represent an important advance for RWE studies that include HF patients. Notably, the only way to ascertain comparative accuracy between data sources and technologies in a domain is to test it. Accuracy consists of both recall and precision, and in the case of many health conditions, recall can fall below 50% when one relies solely on the problem list.31 32
In the current study, use of the F1 score enabled analysis of both precision and recall. Despite availability of SNOMED codes for HFrEF and HFpEF, along with a similar code for HFmrEF, such codes were rarely included. Documentation of a HF code using structured data was only found 46.9% of the time when there was clear evidence of HF in the chart. The low accuracy of structured data for disease subtypes may, at least partially, relate to how the data are likely to be used. A physician may look within notes to understand HF subtype. Information entered into problem lists and claims may be more to provide a high-level understanding of disease burden. Granular billing codes may be a low priority for physicians if claims are reimbursed with the non-granular HF code. Furthermore, because addition of diagnoses to the problem list is not a requirement, the problem list may not be specific or updated. This contrasts with clinical notes, where detailed documentation is usually performed to communicate a care plan and is a medical-legal requirement.
When low-accuracy and non-granular data are utilised, there are several potential consequences. Missingness can result in selection bias, particularly if sicker patients have more frequent encounters, higher rates of specialty care and more complete documentation. Depending on the study question, use of structured data alone to identify certain subgroups may be inadvisable, since these data have a low recall for specific clinical concepts such as ST-elevation myocardial infarction and HFrEF.33 Even advanced approaches (eg, NLP) may result in poor accuracy, as illustrated in this study, where HFpEF required AI-based inference for proper identification. Collectively, this highlights that not all data sources and technologies are the same; therefore, accuracy testing may be required for rigorous RWE generation.34 Furthermore, given the growth in RWE to support new drug indications, postmarketing surveillance, and decision-making regarding reimbursement, it is imperative for clinicians to understand that such inaccuracies may have a profound impact on large numbers of patients.
Even though standard dictionaries and clinical terms related to cardiovascular medicine were used, there is a need to test the two analytic methods using different EHRs across a broader set of community and referral practices. With numerous EHRs available and practitioner-to-practitioner variability in documentation accuracy, efforts like the one described here represent an important means of strengthening data quality.
Importantly, this study has several limitations. First, data from a single health system was used and results may not be generalisable to other populations. Second, the study protocol required the selection of patients enriched with cardiovascular disease to make the study feasible, with manual chart abstraction conducted to ensure the accuracy of results. While selection criteria were applied to both structured and unstructured data, it is possible that this could have biased results in a way that favoured structured data since a larger proportion of patients with HF on the problem list may have been included than if the sample had been created randomly. In addition, the specific filters used likely led to a higher-than-expected proportion of patients with HFrEF (compared with those with HFmrEF and HFpEF). Second, the study required laborious manual annotation of thousands of records. Such a sample size is adequate for high-prevalence conditions, but would likely require adjustment for low-prevalence conditions with low concept occurrence rates. Finally, the study did not include clinical outcome assessment; rather, it was designed to compare data sources and processing methods.
Conclusion
As RWE is increasingly used to analyse patient subgroups, inform clinical decision-making and influence regulatory and reimbursement decisions, data reliability and evidence validity are of critical importance. Use of a traditional approach was associated with low data accuracy. While much greater accuracy was observed with AI-based methods, it depended on the technology utilised. These findings highlight the importance of using data fit-for-purpose to the research question posed. In addition, they suggest that accuracy testing should be part of any EHR-based study that includes patients with HF. Finally, unstructured data and a technology-based approach to data extraction may be required in some studies to achieve sufficient accuracy, depending on the clinical assertion being tested.
Data availability statement
No data are available.
Ethics statements
Patient consent for publication
Ethics approval
This study has been independently reviewed and accepted for exemption in accordance with 45 CFR 46.101(b)(4ii).
Acknowledgments
We are grateful for comments from Jacob Abraham, MD, Medical Director at Providence Heart Institute's Center for Advanced Heart Disease, and Yuri Quintana, MD, Chief of, Division of Clinical Informatics at the Beth Israel Deaconess Medical Center. Editorial support was provided by Liam Gillies, PhD, CMPP, of Cactus Life Sciences (part of Cactus Communications), funded by Amgen Inc.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
Correction notice This article has been corrected since it was first published. The funding statement has been updated.
Contributors ARG and DJR drafted the manuscript. ARG, KLM, RED-A, DJR and TJG critically reviewed the manuscript. ARG, RED-A, DJR and TJG provided clinical insight. DJR is the study guarantor.
Funding A research grant supported this work from Amgen Inc. DJR was partly supported by the US Food and Drug Administration (FDA) under Award Number U01FD007172 and the National Center for Advancing Translational Sciences of the NIH under Award Number R44TR002437. The content is solely the responsibility of the authors and does not necessarily represent the official views of Amgen, the FDA or the NIH.
Competing interests KLM and RED-A are employees and stockholders of Amgen Inc. DJR is an employee and stockholder of Verantos, Inc. ARG has received research support from Abbott.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting or dissemination plans of this research.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.